Key feature
Automated muscle quantification
MuscleMap's `mm_extract_metrics` is the command-line tool for quantitative muscle analysis, enabling automated estimation of muscle volume, cross-sectional area (CSA), intramuscular fat infiltration, and muscle density from MRI and CT images.
It supports Gaussian Mixture Models (GMM), K-means clustering for T1 and T2-weighted MRI, fat fraction for Dixon MRI and HU‑based fat quantification for CT in customizable (.csv) output formats.
This page explains:
- how
mm_extract_metricsworks - all key command-line options
- recommended workflows
- output files and their interpretation
- troubleshooting guidance
Tip: For the most up-to-date options in your installed version, run:
mm_extract_metrics --help
1. Basic usage
After generating a segmentation with mm_segment, run:
mm_extract_metrics -m gmm -r wholebody -i image.nii.gz -s image_dseg.nii.gz -c 3
This command:
- loads the image (
-i) - loads the segmentation (
-s) - applies the chosen metric method (
-m) - computes fat/water composition or tissue-specific statistics
- outputs CSV + NIfTI metric maps (depending on options)
2. Required inputs
2.1 -i — Input image
The MRI/CT image from which metrics are extracted:
mm_extract_metrics -i sub-01_T2w.nii.gz
2.2 -s — Muscle segmentation labelmap
The muscle segmentation labelmap produced by mm_segment or from manual segmentation:
mm_extract_metrics -s sub-01_dseg.nii.gz
The segmentation must contain the same dimensions and orientation as the input image.
2.3 -o — Output directory
Output directory to save the results from mm_extract_metrics. If not specified, the results are saved in the same directory as the input image.
3. Metric computation methods (-m)
The -m flag determines how fat fraction / composition metrics are computed.
Supported values:
1. Dixon — Fat–water–based metrics
Uses Dixon-based fat and water separation to compute voxel-wise fat fraction and derive muscle composition metrics within the muscle segmentation.
mm_extract_metrics -m dixon -f fat.nii.gz -w water.nii.gz -s img_dseg.nii.gz
2. gmm — Gaussian Mixture Model (MRI)
Uses a Gaussian Mixture Model (GMM) to separate tissue types by fitting multiple Gaussian distributions to the intensity histogram and classifying voxels based on their intensity-derived probabilities.
mm_extract_metrics -m gmm -i img.nii.gz -s img_dseg.nii.gz
3. kmeans — Kmeans clustering (MRI)
Uses k-means clustering to partition voxels into intensity-based clusters by minimizing within-cluster variance, assigning each voxel to the nearest cluster centroid (e.g., fat vs. muscle) based on its intensity.
mm_extract_metrics -m kmeans -i img.nii.gz -s img_dseg.nii.gz
Use GMM and/or K-means clustering on T1-weighted or T2-weighted MRI only.
4. average — Density metrics
An averaging-based method to quantify muscle density by computing the mean voxel signal intensity within the muscle region..
mm_extract_metrics -m average -i ct_img.nii.gz -s ct_dseg.nii.gz
Use `average` metrics preferably for CT. MRI intensities do not reflect physical density.
4. Region selection (-r)
Choose which muscle regions to extract metrics for.
Example:
mm_extract_metrics -r wholebody
Common values include:
wholebody(default)abdomenpelvisthighleg
Regions correspond to MuscleMap’s anatomical label groups.
Region definitions are based on the MuscleMap atlas and segmentation model.
5. Number of clusters (-c)
Used only with GMM or Kmeans and T1- or T2-weighted MRI.
Example for 3 clusters:
mm_extract_metrics -c 3
Typical choice:
- 2 clusters: fat vs. muscle
- 3 clusters: fat, muscle, intermediate tissue
Intermediate reflects a voxel with intermediate voxel signal not clearly corresponding to either fat or muscle.
6. Output files
mm_extract_metrics typically produces:
1. CSV file with summary statistics
Contains per-muscle metrics, dependent on the arguments, such as:
- muscle volume
- fat fraction
- average density (e.g., for CT)
2. Voxel-wise metric maps (optional depending on method)
Examples:
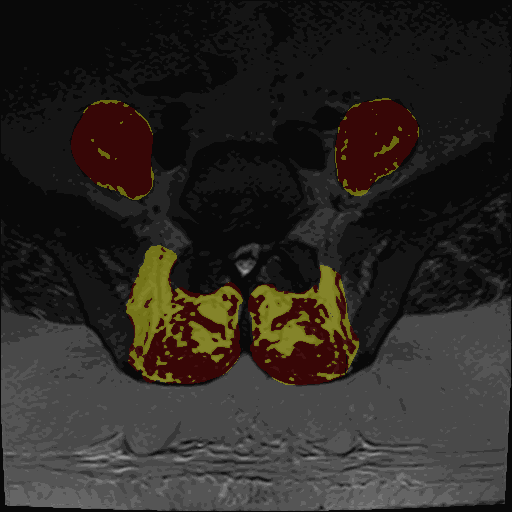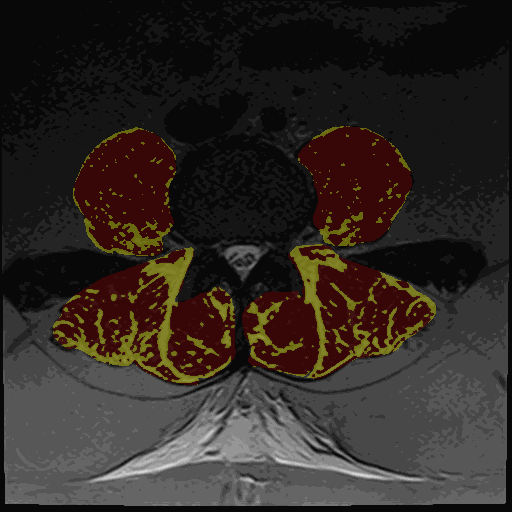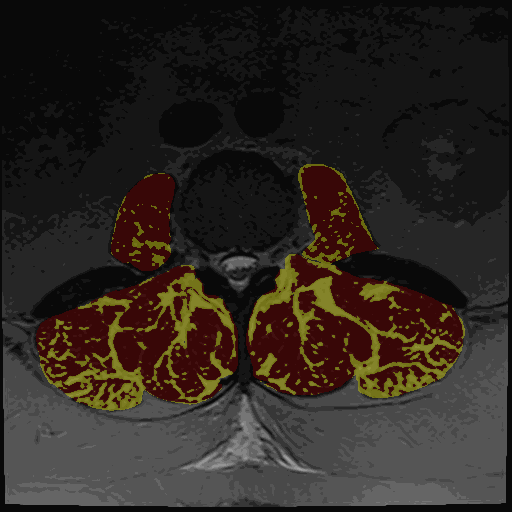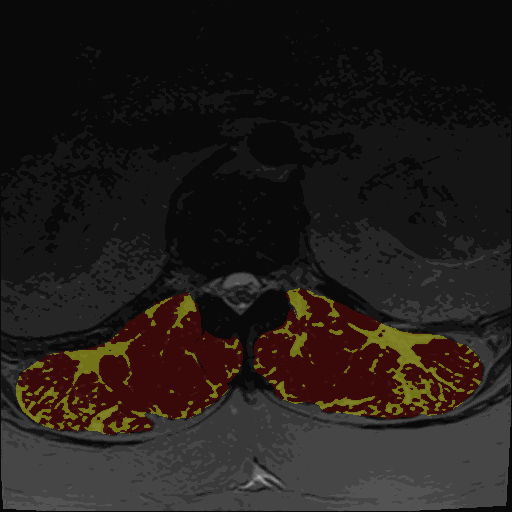
- Thresholding maps for either GMM or Kmeans and two or three clusters
The thresholding maps can be loaded as a segmentation to visually check thresholding accuracy
7. Example workflows
7.1 MRI (T1/T2) using GMM
mm_segment -i sub-01_T2w.nii.gz
mm_extract_metrics -m gmm -i sub-01_T2w.nii.gz -s sub-01_T2w_dseg.nii.gz -r wholebody -c 3
7.2 MRI using k-means
mm_extract_metrics -m kmeans -i img.nii.gz -s img_dseg.nii.gz -r pelvis
7.3 CT muscle density (HU)
mm_extract_metrics -m hu -i ct_img.nii.gz -s ct_dseg.nii.gz -r abdomen
8. Best practices & troubleshooting
Always **visually inspect both segmentation and metric outputs** before analysis.
9. Summary
mm_extract_metrics is the quantitative analysis backbone of MuscleMap:
- accepts MRI or CT images + segmentations
- computes fat fraction, HU metrics, tissue composition
- supports GMM, Kmeans, HU-based methods
- outputs CSV and optional maps
- integrates with
mm_guifor streamlined workflows