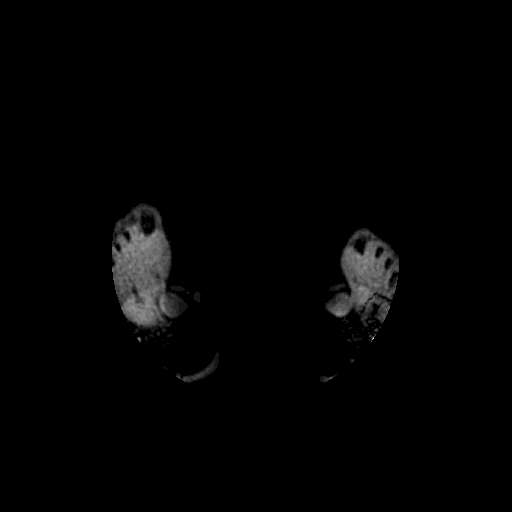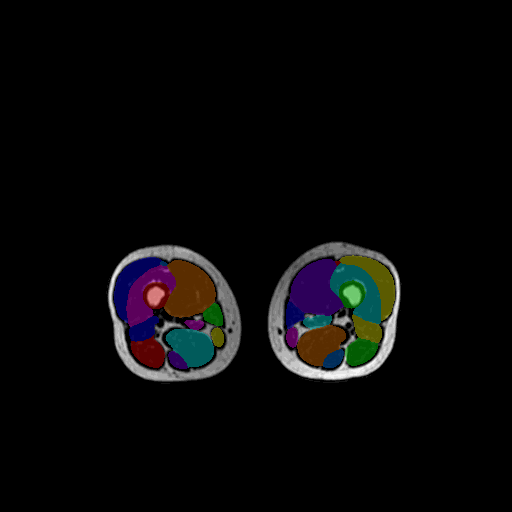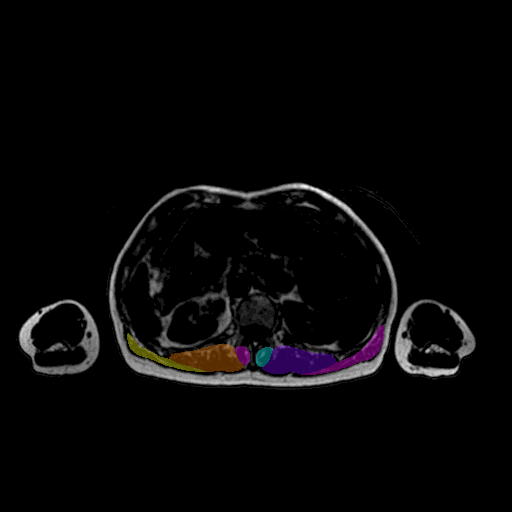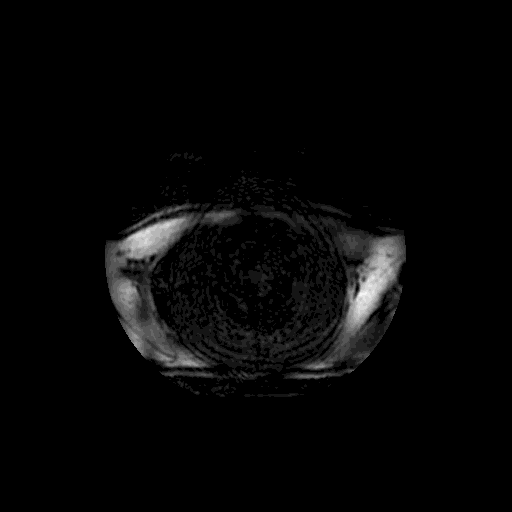
Key feature
Automated whole-body muscle segmentation
MuscleMap segmentation is performed using a deep learning–based, contrast-agnostic whole-body model trained for robust muscle and bone segmentation across MRI and CT modalities.
This page explains:
- how to run segmentation module:
mm_segment - all available options and flags
- whole-body vs. regional models
- recommended workflows
- troubleshooting tips
Tip: For the most up-to-date list of options in your installation, run:
mm_segment --help
1. Basic usage
1.1 Activate the MuscleMap environment
conda activate MuscleMap
1.2 Run mm_segment on a single NIfTI image
First, navigate to the directory containing your input image (or provide the full path to the file):
cd /path/to/your/data
Then run:
mm_segment -i image.nii.gz
This command:
- loads the input image
- applies the default whole-body segmentation model
- writes a segmentation labelmap (NIfTI) next to the input file
- prints output paths and logs to the terminal
Running
mm_segment from the command line works as expected when MuscleMap is installed in editable mode (pip install -e .).If the package was installed using
pip install ., it may be preferable to run the script directly using python mm_segment.py. 2. Input requirements
2.1 Supported image types
- Modality: Axial MRI of any contrast (T1w, T2w, Dixon water/fat/in-phase) or CT
- Format: NIfTI (
.nii,.nii.gz) - Orientation: Axial orientation recommended
MuscleMap recommends storing data in BIDS format with JSON sidecars from dcm2niix.
2.2 Processing single images vs. datasets
mm_segment processes one image at a time.
For multiple images:
- loop through images in shell/Python
- keep consistent naming using BIDS
3. Output
mm_segment generates deep learning–based whole-body muscle segmentation outputs, including:
- a segmentation labelmap (NIfTI) with integer labels
- an output file saved next to the input
- terminal logs with paths and runtime info
Visualise results in:
4. Models and regions
4.1 Whole-body model (default)
mm_segment -i image.nii.gz
Segments:
- trunk muscles
- pelvic muscles
- thigh muscles
- leg muscles
- neck muscles
- selected bones
4.2 Legacy regional models
Available:
abdomenpelvisthighleg
Example:
mm_segment -i image.nii.gz -r abdomen
The legacy regional models are maintained for backward compatibility only. Active development and state-of-the-art performance are provided exclusively by the whole-body model, which achieves robust performance across all anatomical regions.
5. Command-line options
5.1 -i — input image (required)
mm_segment -i /path/to/image.nii.gz
5.2 -s — sliding-window overlap
Controls tile overlap.
Example:
mm_segment -i image.nii.gz -s 50
Lower sliding-window overlap increases inference speed but may reduce segmentation performance, particularly near tile boundaries. Higher overlap improves robustness at the cost of longer runtimes.
5.3 -r — region model
mm_segment -i image.nii.gz -r thigh
5.4 -g — GPU/CPU selection
Controls whether inference is performed on the GPU or CPU.
Example:
mm_segment -i image.nii.gz -g Y
By default, MuscleMap runs inference on the **GPU** when available. Use `-g N` to explicitly force CPU-based inference.
5.5 -h — help
mm_segment -h
6. Example workflows
6.1 Whole-body segmentation
mm_segment -i sub-01_water.nii.gz
6.2 Faster inference
mm_segment -i sub-02_T2w.nii.gz -s 50
6.3 Using a regional model
mm_segment -i sub-03_T2w.nii.gz -r abdomen
6.4 Full pipeline with metrics
mm_segment -i image.nii.gz
mm_extract_metrics -m gmm -r wholebody -i image.nii.gz -s image_dseg.nii.gz -c 3
See the Muscle quantification and metric extraction page for details on muscle volume and fat infiltration analysis.
7. Best practices & troubleshooting
- Always visually inspect segmentations
- Poor image quality reduces accuracy
- Record MuscleMap version and options in publications
If something looks wrong:
- open a GitHub issue with an example