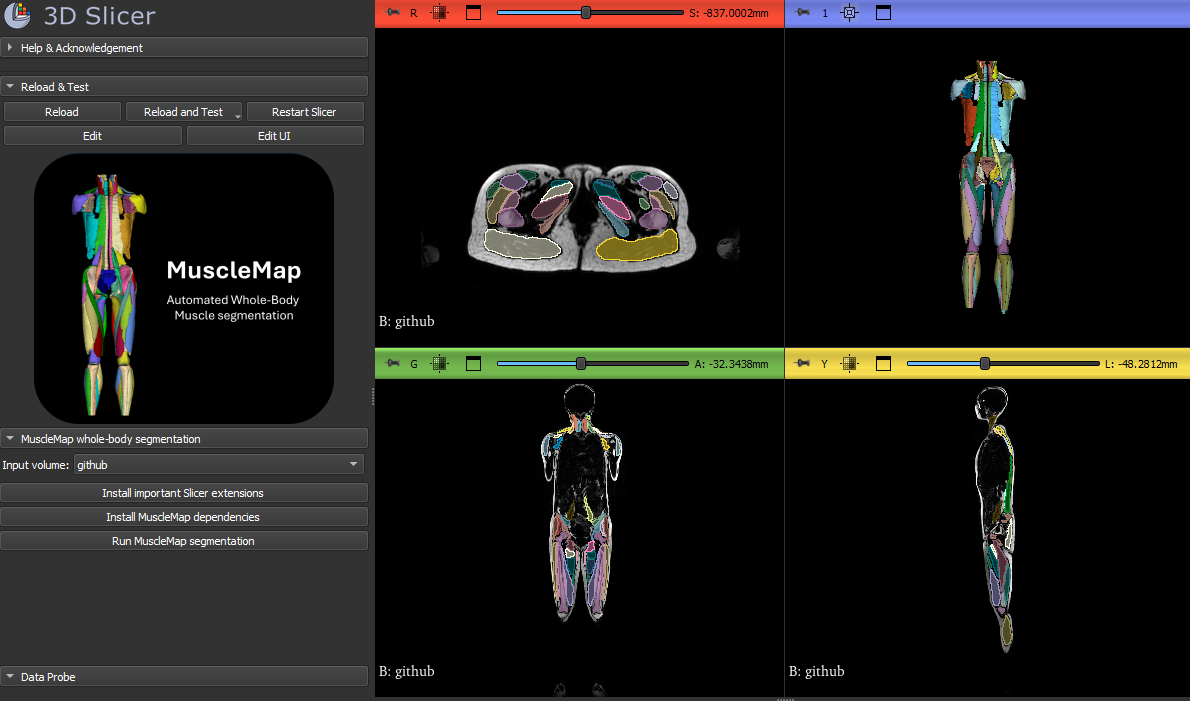
3D Slicer extension for muscle segmentation
MuscleMap is implemented as a dedicated 3D Slicer extension that enables fully automated whole-body muscle segmentation directly within the Slicer environment. This interface is designed for researchers, clinicians, and imaging scientists who want to apply MuscleMap models without writing code.
The GUI provides an end-to-end workflow: from loading CT or MRI data, installing dependencies, running segmentation, to interactive 3D visualisation of muscle labels.
Why use the MuscleMap 3D Slicer extension?
- No programming required
- Fully integrated into 3D Slicer
- Supports CT and MRI
- Automated whole-body muscle segmentation
- Interactive 2D and 3D visualisation
- Export-ready segmentation labels for quantitative analysis
Installation
Step 1 – Install 3D Slicer
Download and install the latest version of 3D Slicer (≥ 5.2) from:
https://www.slicer.org
Step 2 – Install the PyTorch (SlicerPyTorch) extension
MuscleMap requires PyTorch to run deep-learning models inside Slicer.
- Open 3D Slicer
- Go to:
View → Extensions Manager - Search for:
PyTorch or SlicerPyTorch - Install the extension
- Restart 3D Slicer
Step 3 – Install PyTorch inside Slicer
After restarting Slicer:
- Go to:
View → Modules - Navigate to:
Utilities → PyTorch (SlicerPyTorch) - Click:
Install PyTorch - Choose:
- GPU version (recommended if a compatible GPU is available)
- or CPU version otherwise
Wait until the installation is completed successfully.
Step 4 – Install the MuscleMap extension
- Open Extensions Manager again
- Search for:
MuscleMap - Install the extension
- Restart 3D Slicer
After restarting, the module is available under the MuscleMap category.
Using SlicerMuscleMap
1. Open the MuscleMap module
Go to:
View → Modules → MuscleMap → MuscleMap Whole-Body Segmentation
2. Install MuscleMap dependencies (one-time step)
Inside the module:
- Open Installing packages
- Click:
Install MuscleMap dependencies
This installs MONAI, nibabel, pandas, and the MuscleMap toolbox inside Slicer’s Python environment.
3. Load an image volume
You can load data in two ways:
- Click Load volume from file…
- Or select an already loaded volume using Input volume
Supported formats include:
.nii,.nii.gz
Both CT and MRI are supported.
4. Run MuscleMap segmentation
- Select the input volume
- (Optional) Enable Force CPU under Advanced if needed
- Click:
Run MuscleMap segmentation
The model will automatically:
- Export the volume
- Run
mm_segment - Load the resulting segmentation
- Apply anatomical labels and colors
- Display the result in 2D and 3D
5. Visualise results in 3D
After segmentation:
- The output segmentation appears automatically
- Click Show 3D to enable 3D rendering
- Use standard Slicer tools for inspection, annotations or export
Source code and development
The 3D Slicer extension is open-source and actively developed.
Reporting issues and requesting features
If you encounter an error, unexpected behaviour, or have a feature request, please open an issue on GitHub.
Providing the following information helps us resolve issues faster:
- 3D Slicer version
- Operating system
- CPU/GPU details
- Error message or log output
- Example data (if possible)
Citation
If you use MuscleMap for academic or scientific work, please cite the MuscleMap consortium publications listed in the module acknowledgements.
MuscleMap – Open-source, community-supported whole-body muscle quantification.